Investigating the microbial basis of population suppression in Australian plague locusts
Cases of Australian plague locust population suppression occurred despite extreme swarming conditions forecasted for the wet Australian summer. Investigations into cause of this population suppression could lead to novel ways to protect cereal crops from this pest species.
Challenge

Healthy Australian plague locust.

Diseased Australian plague locust.
Locusts are typically solitary insects, however, when environmental conditions are favourable, their behaviour alters to form swarms (known as a gregarious phase). High-density swarms of locusts, known as plagues, feed voraciously on plants and can devastate large areas of agricultural land. During plague years, the Australian plague locust (APL) is Australia’s most serious pest species, causing A$25 million in damages to cereal crops (agriculture.gov.au). Extreme swarming events were forecast for the wet Australian summers of 2020-2022. However, locusts remained solitary and population numbers remained low. Despite this favourable outcome, the Australian Plague Locust Commission wished to investigate the causes of this population suppression with the anticipation of finding novel management solutions for this pest species and the protection of cereal crops.
Our response
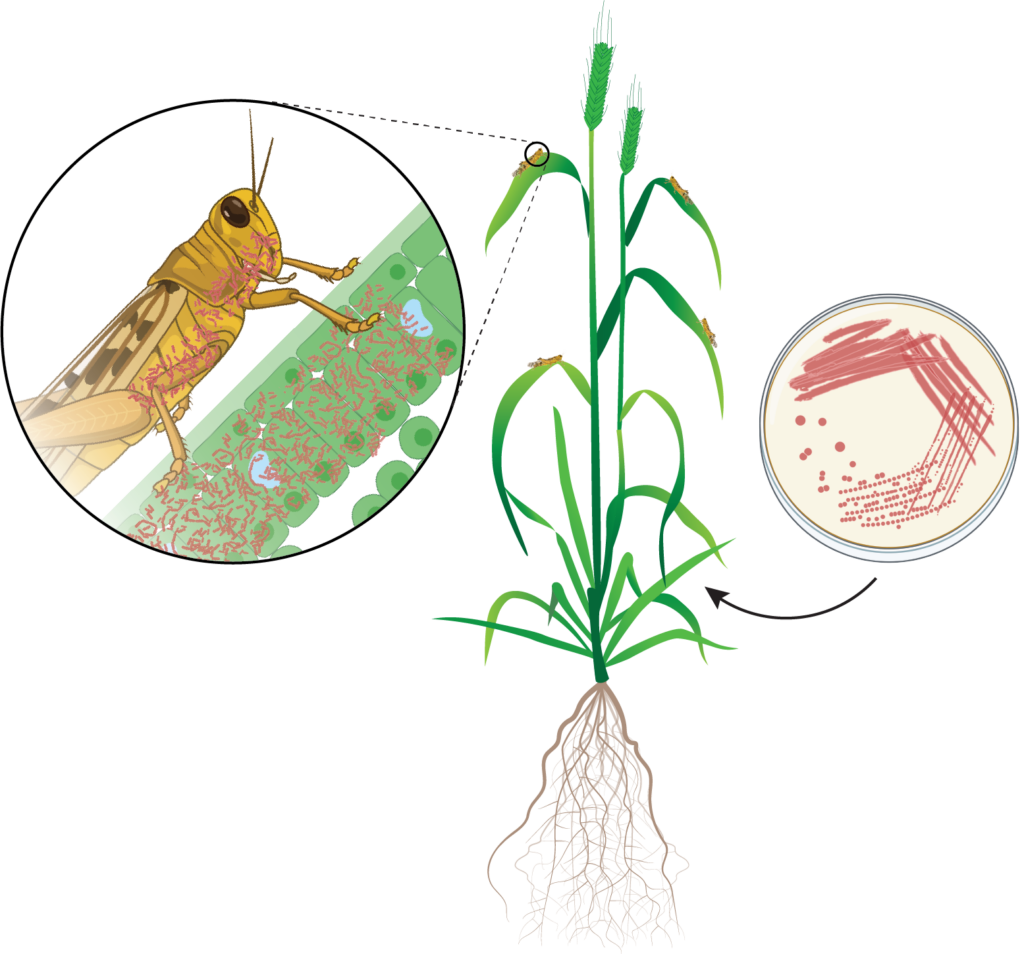
A covert insect pathogen was a strong candidate for this observed population suppression, and the observation of locusts with red pigmentation in affected populations only strengthened this theory. The aim of the study was to identify a pathogen suspected of suppressing specific populations of APL. To identify the putative pathogen, 16S and ITS amplicon sequencing was performed, creating a molecular census of all the bacteria and fungi present in the healthy and diseased populations of locusts. This strategy involves isolating all the microbial DNA from the sample locusts and amplifying a highly conserved gene found in all microbial genomes. Using high-throughput sequencing, subtle differences in the genetic code of these genes are then used to distinguish which microbes are present and even provide approximations of their abundance in the sample.
Locust endophyte cycle
One bacterial group stood out as a strong contender; despite only being present in samples collected from suppressed populations, it had the highest relative abundance of all bacterial species identified in the entire study. Species from this bacterial group are found in soil, saltwater, and freshwater and are known to variously infect insects, plants and animals.
We will be performing targeted culturing of strains-of-interest and infection-based assays of living locusts to determine a causative link between this pathogen and locust population suppression. Once established, further detailed genomic and molecular analyses will be used to identify potential targets that could be used as a novel biopesticide. Further, by understanding the genetics underpinning solitary-to-gregarious phase shifts could be used as a potential target for management solutions of this pest species.
Our team
Dr Simon Law – Research scientist
Dr Luke Barrett – Research scientist
Moazzem Khan – Collaborator with the Australian Plague Locust Commission (funding body)
Find out more at the Australian Plague Locust Commission – DAFF (agriculture.gov.au)
