BLAST against the NBDL
You can search your own DNA sequences against the full NBDL database using the BLAST algorithm. BLAST stands for Basic Local Alignment Search Tool, and it is one of the most common algorithms used to compare DNA sequences. BLAST performs a local sequence alignment of a query sequence against a database of reference sequences, returning the closest matches in the database to the submitted query. When you query your own DNA sequence against the NBDL, it will be compared against all the available sequences from Australian collection specimens. The version of BLAST used in the NBDL is currently 2.16.0+.
To query a DNA sequence, click on the Query option on the main menu or click the Query button below the Search bar on the main page. This will take you to the BLAST search page (see image below).
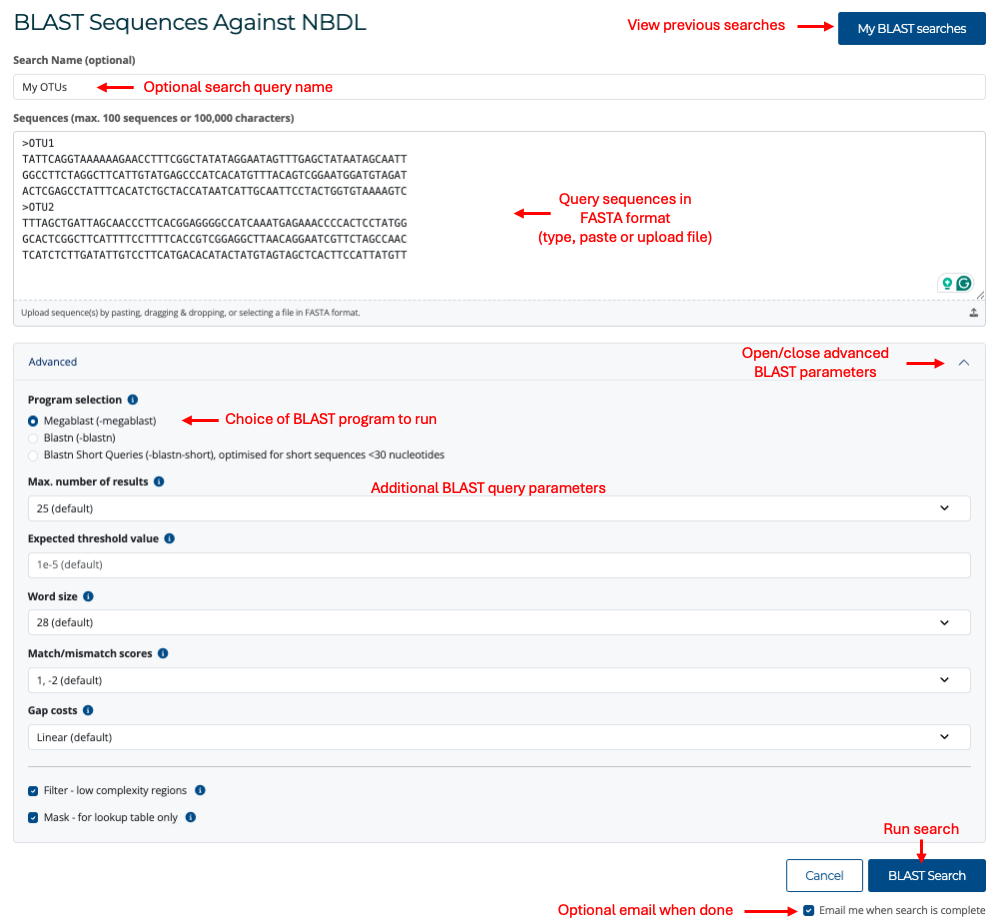
In the BLAST query page, you can type or paste DNA sequences directly or choose to upload a file. Sequences must be in FASTA format. More information on this format can be found here. You can optionally enter a search query name for your BLAST search. All BLAST searches are saved for 30 days, and you can use this name to identify your BLAST search from the list of previous searches.
In the advanced parameters menu, you can select the most appropriate BLAST search parameters for your query. There are three program options to choose from:
- Megablast is intended for comparing a query to closely related sequences and works best if the target percent identity is 95% or more, but it is very fast.
- Blastn is slow but allows a word size down to seven bases.
- Blastn-short is optimised for short sequences of less than 30 nucleotides.
Each of these options will adjust the appropriate parameters based on the defaults for each program. You can further change these parameters to suit your search. If you ignore the advanced parameters menu, the default option will use MegaBlast. For more information on how BLAST programs work, see the BLAST help page here.
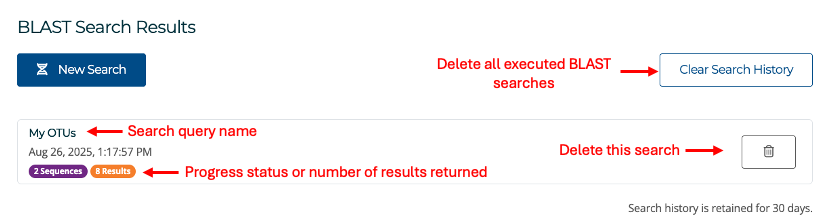
Once your search is submitted, you will be redirected to the BLAST search results page, which will list your current and previously executed BLAST searches (see image below). Each search result row shows the name of the search query, the date and time it was submitted and the current progress status of the query (if not completed). You can delete any search from your list of searches by clicking the delete button at the end of the row or you can delete all previous BLAST searches by clicking on the Clear Search History button.
Clicking on the search name or the row itself will take you to the BLAST search results page (see image below). Within this window you can view the matching sequences for each of your individual query sequences by navigating through the results using the navigation button above and below the list of results. Each result will show the species name and specimen ID matched to the query, and the identity score of the match. The matches will be sorted by lowest expected value (E value). For more details on how these scores are calculated, see BLAST FAQs. To view the aligned matching region, you can click on the arrow at the end of each result. This will show the matching source reference sequence and the FASTA header. The alignment section displays your query sequence at the top and the matching region in the reference at the bottom.
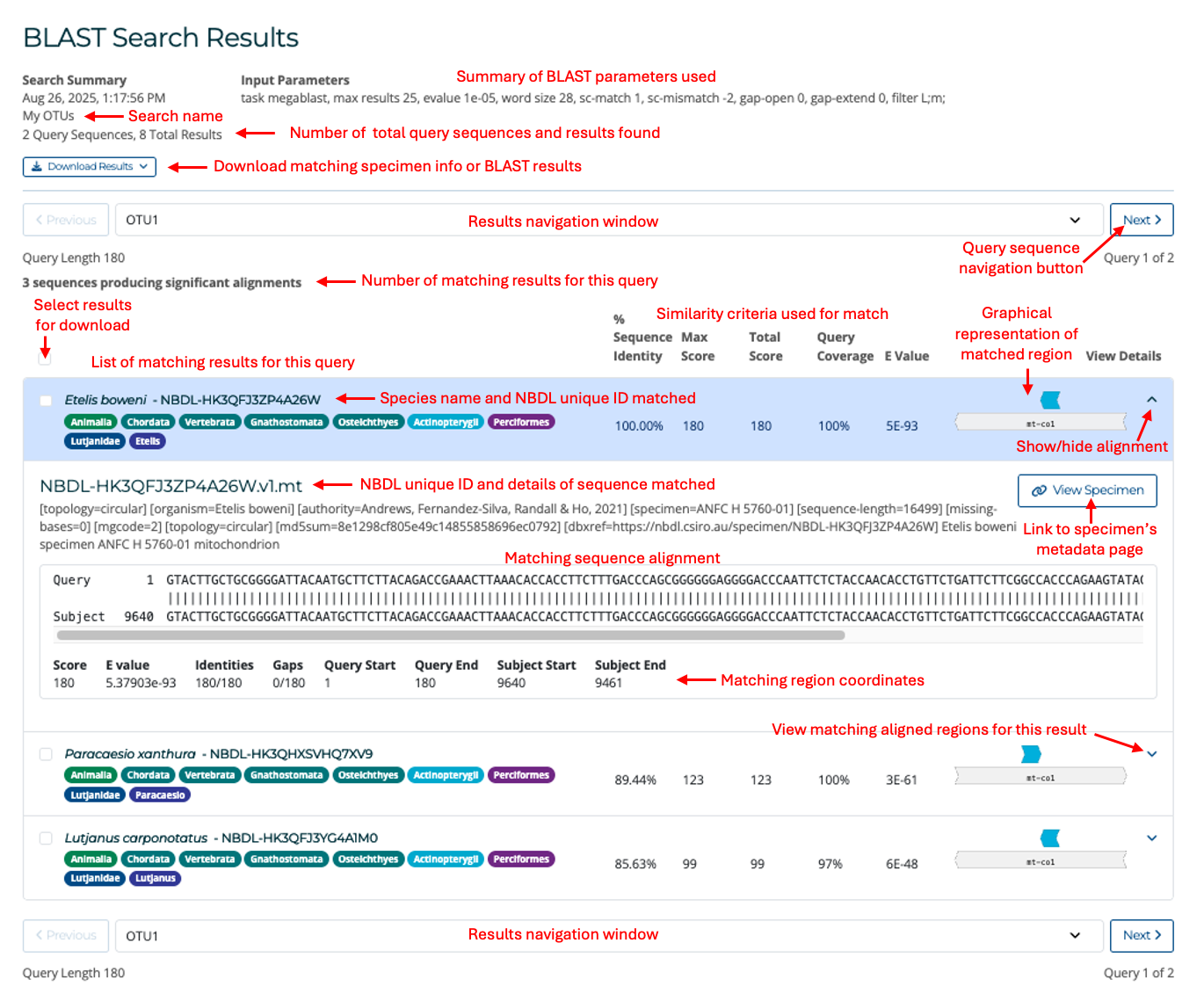
To download BLAST results or matching specimen data, click on the Download Results button. From the Download Results menu, the following options are available:
- Download selected: Download a selection of matching specimen sequences. Before downloading, you can select individual specimen sequences to download by clicking on the selection buttons on the left of each result in the BLAST Search Results screen.
- Download all: Download all matching specimen sequences.
- Standard tabular report, TSV: Download a tab-separated text file containing a summary of matching BLAST hits to query sequences. Contains one result per line displaying specimen ID, similarity scores and aligned region coordinates.
- Full tabular report, TSV: Download a tab-separated text file containing the above fields plus additional information such as the aligned regions and specimen sequence headers.
- Full XML report, XML: Same as the full tabular report above, but in XML format.
- Clear selection:Clear the specimen selections made in the BLAST Search Results screen.
If you select either of the first two download options, you will be taken to the Download data dialog to choose the sequence regions for download. For more information on the download options available, go to Download data.
